By Georgia Barrington-Smith & Dr Rebecca Duncan
Ensuring our ongoing food availability in the face of a rising global population is a critical challenge. Infectious plant diseases pose a significant threat to our agricultural food production, costing the global economy around $220 billion USD each year.
One particularly destructive disease is ‘blast disease’, which targets valuable cereal crops like rice, wheat, and barley, leading to substantial global food losses. Research focusing on new methods to prevent plant diseases, like blast disease, has never been more vital.
Those pesky pathogenic fungi!
Blast disease is caused by the fungus Magnaporthe oryzae, a pathogenic microbe that secretes small proteins called ‘effectors’ into the host plant, causing infection and disease. As little is currently understood about how these effectors function, AINSE PGRA scholar Carl McCombe, together with his collaborators at ANSTO and the Australian National University, set out to address these critical knowledge gaps.
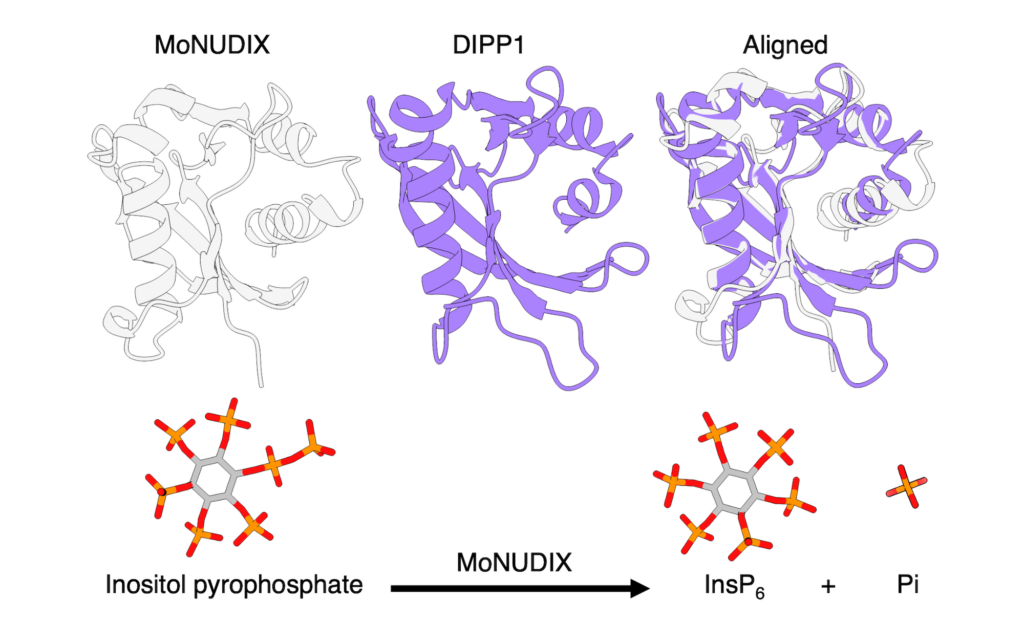
To better understand the mechanisms employed by these pathogenic fungi, Carl used X-ray diffraction measurements on ANSTO’s Australian Synchrotron MX2 beamline to investigate the structure of a particular M. oryzae effector protein, known as MoNUDIX.
Interestingly, the team found that both the structure and function of MoNUDIX were remarkably similar to the human enzyme DIPP1; both can damage the signalling molecules required for regulating phosphate levels inside cells. This disruption to phosphate levels in plant cells activates the Phosphate Starvation Response (PSR), which results in reductions in plant growth, leaf discolouration, and leaf damage.
Are ‘effectors’ affecting all plants?
As many other pathogenic fungi have effectors that function just like MoNUDIX, Carl and his collaborators designed a method to test if these other effectors could also trigger a phosphate starvation response in their host plant in a similar way.
Carl and the team developed a genetic system in which phosphate starvation in leaves generates a visible red pigment. Using this method, the team were able to show that numerous types of pathogenic fungi used a common strategy to make plant diseases worse through the manipulation of phosphate signalling.
By determining the infection mechanisms of plant diseases, Carl and his research team are making vital contributions to a growing field of research focusing on disease resistance crops. These efforts could substantially reduce the devastating food losses caused by fungal pathogens.
AINSE are proud to spotlight Carl McCombe for his breakthrough work!
To read more research spotlights visit ainse.edu.au/research-spotlight.
Keep connected for the second article in our Fungi February series, as we grow our knowledge on pathogenic fungi, and explore PGRA scholar Daniel Vu’s research, looking at plant immune response to fungal effectors.
Stay up to date with AINSE by following us on all our social media platforms @ainse_ltd on Instagram, Facebook, Threads, LinkedIn and X.
